
Theoretical Modeling and Biophysics
We perform theoretical modeling and biophysics involving nanomaterials and biological molecules for example DNA and proteins. The present research is focused on the interaction behavior of materials and biology. These interactions could be understood and analyzed using several techniques, among them theoretical modeling and molecular simulations were used vastly and popular in scientific community.
The software's used for molecular modeling are PyMol, Chimera and VMD etc. For molecular Dynamics simulations, we use NAMD (http://www.ks.uiuc.edu/Research/namd/) offered as open source simulation software from UIUC. The molecular simulations were performed on GPU cluster provided by HKUST (https://itsc.ust.hk/services/academic-teaching-support/high-performance-computing/gpu-cluster/). Our group had recently invested to buy new cluster services with 2XK80 Tesla Nvidia GPU, further it will be expanded.
At present group research focoused on binding energy calculations between biomolecules and materials, the methods used are PMF calculations using ABF, US, MMPBSA, and REUS.




Modelled using VMD
The vedio shows (left side) how DNA and graphene closely interacts during molecular dynamics simulations.
The DNA and graphene have very good interactions, the image (right side) shows how DNA (ssDNA and dsDNA) interacts with graphene. The interactions could be in different orientations.
DNA and Graphene Interactions
Graphene nanopore for DNA sequencing
The vedio shows (left side) how DNA passed through graphene nanopore in the presence of ion. The video on right side shown without ions for clear understanding.
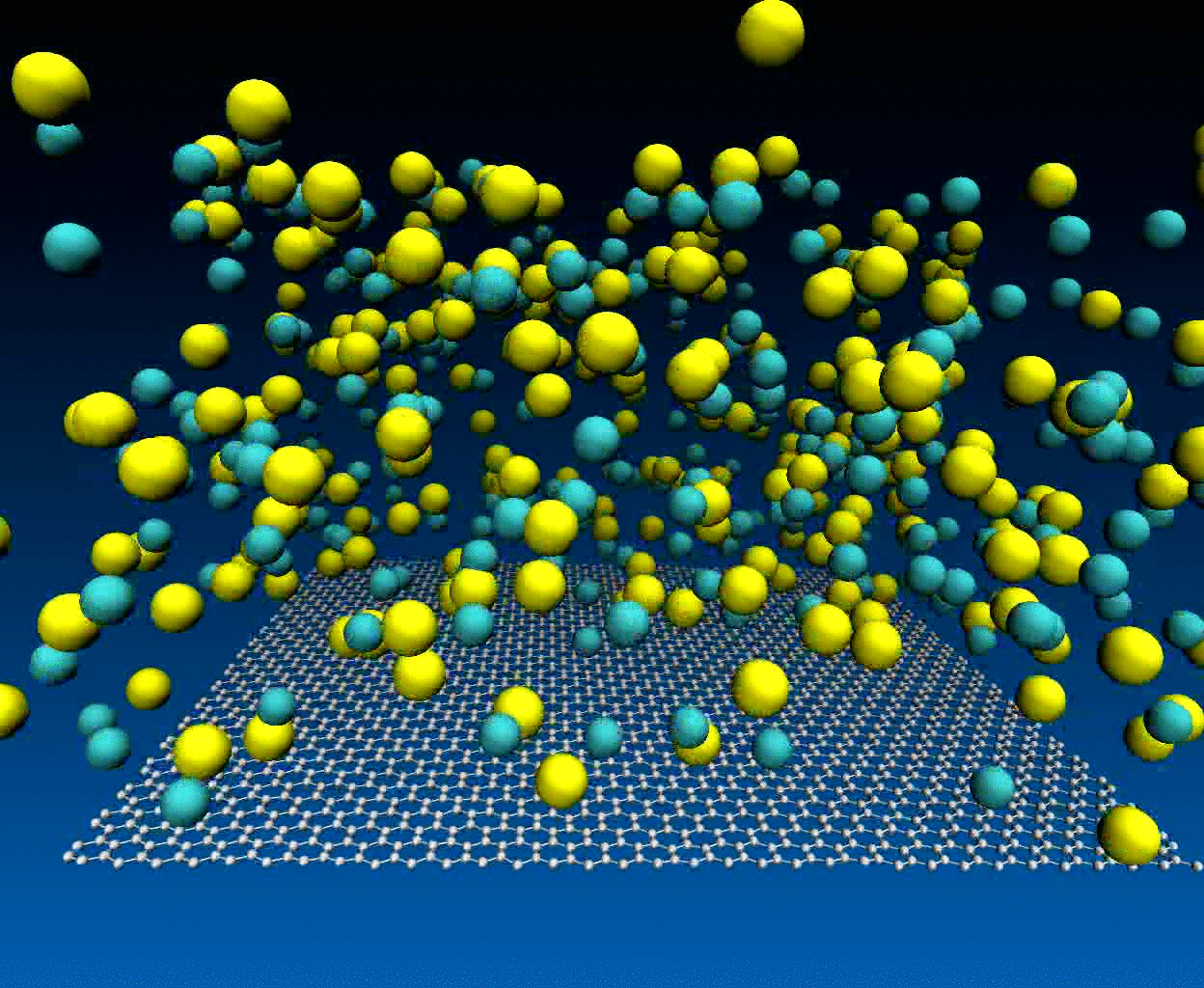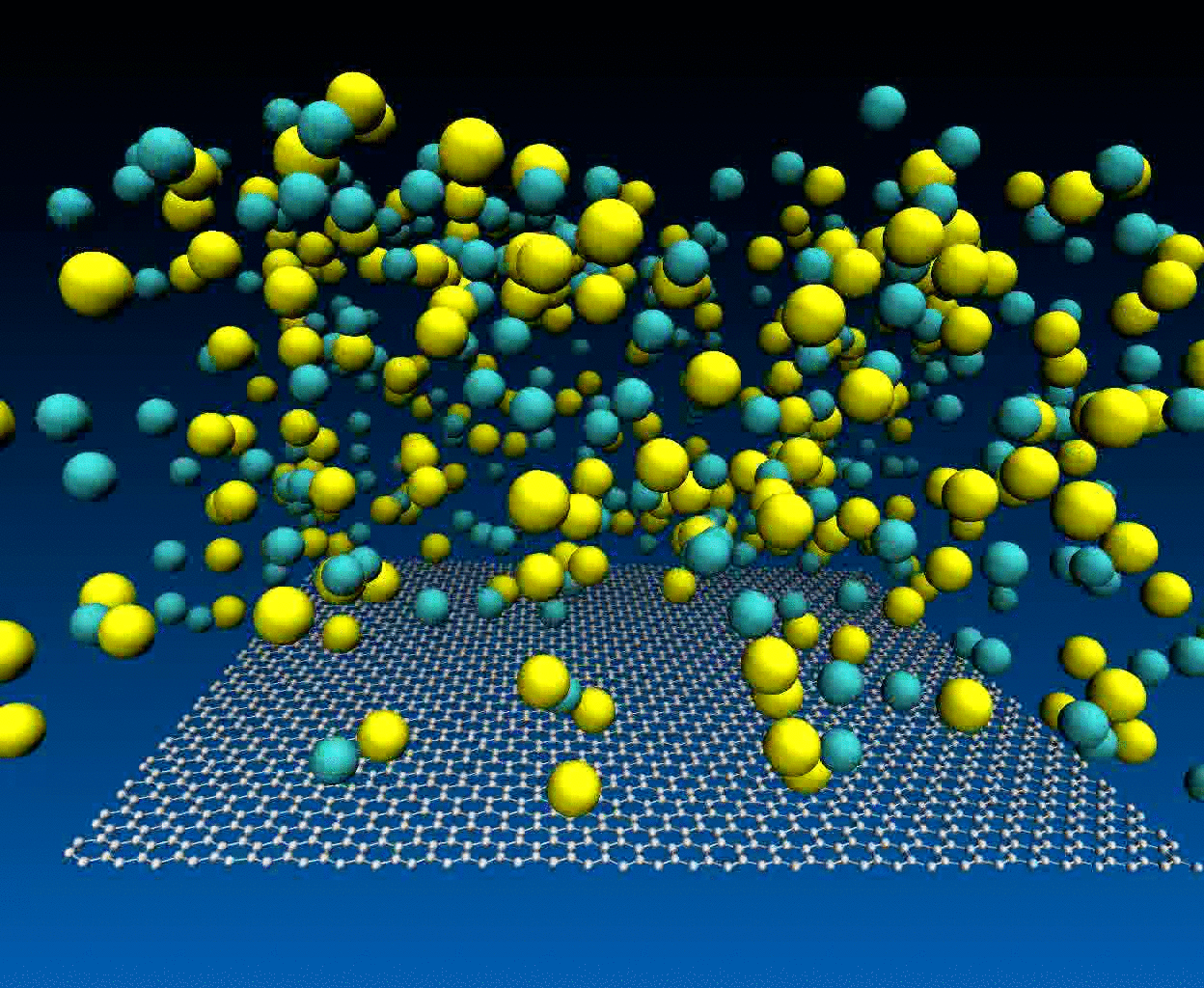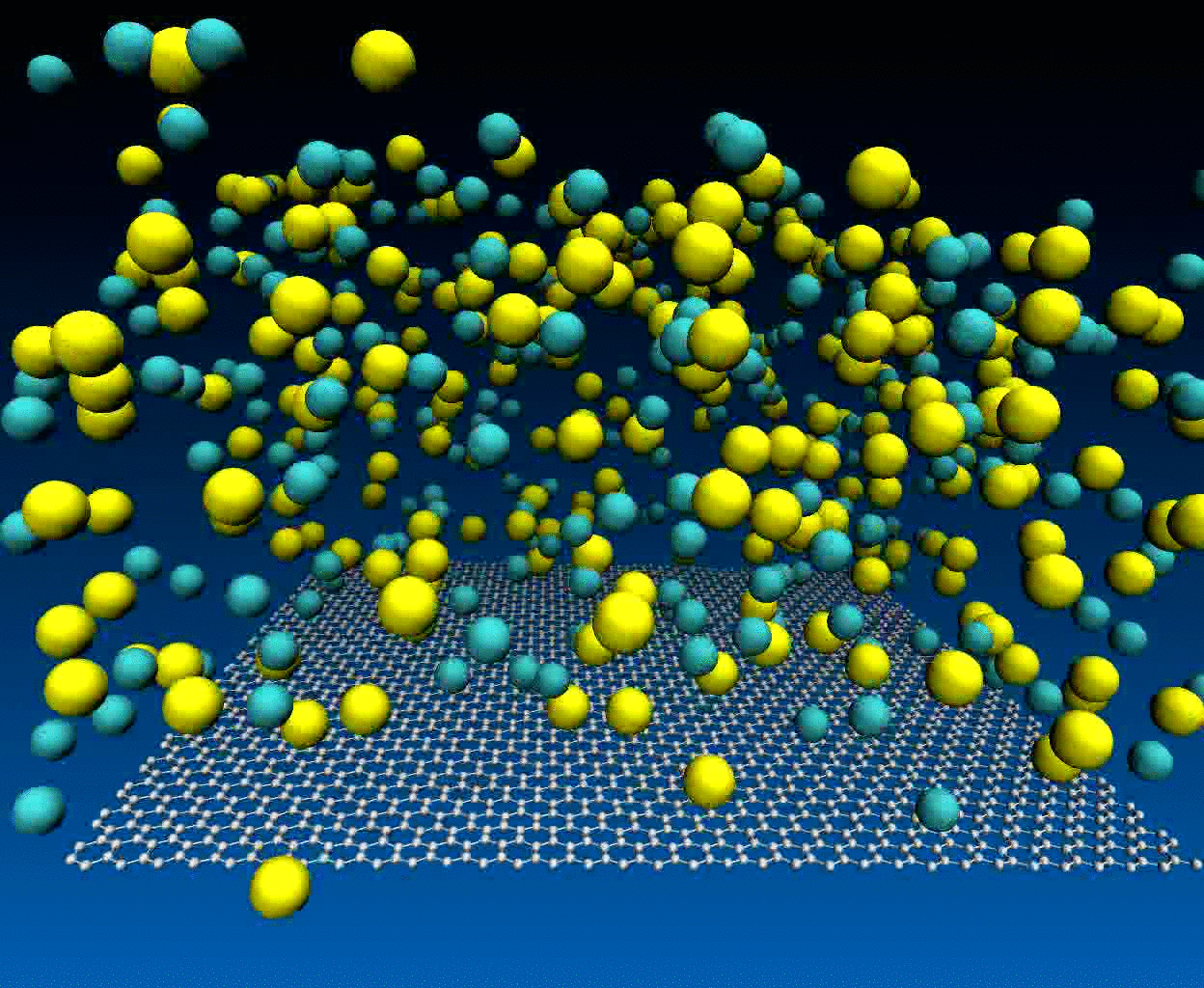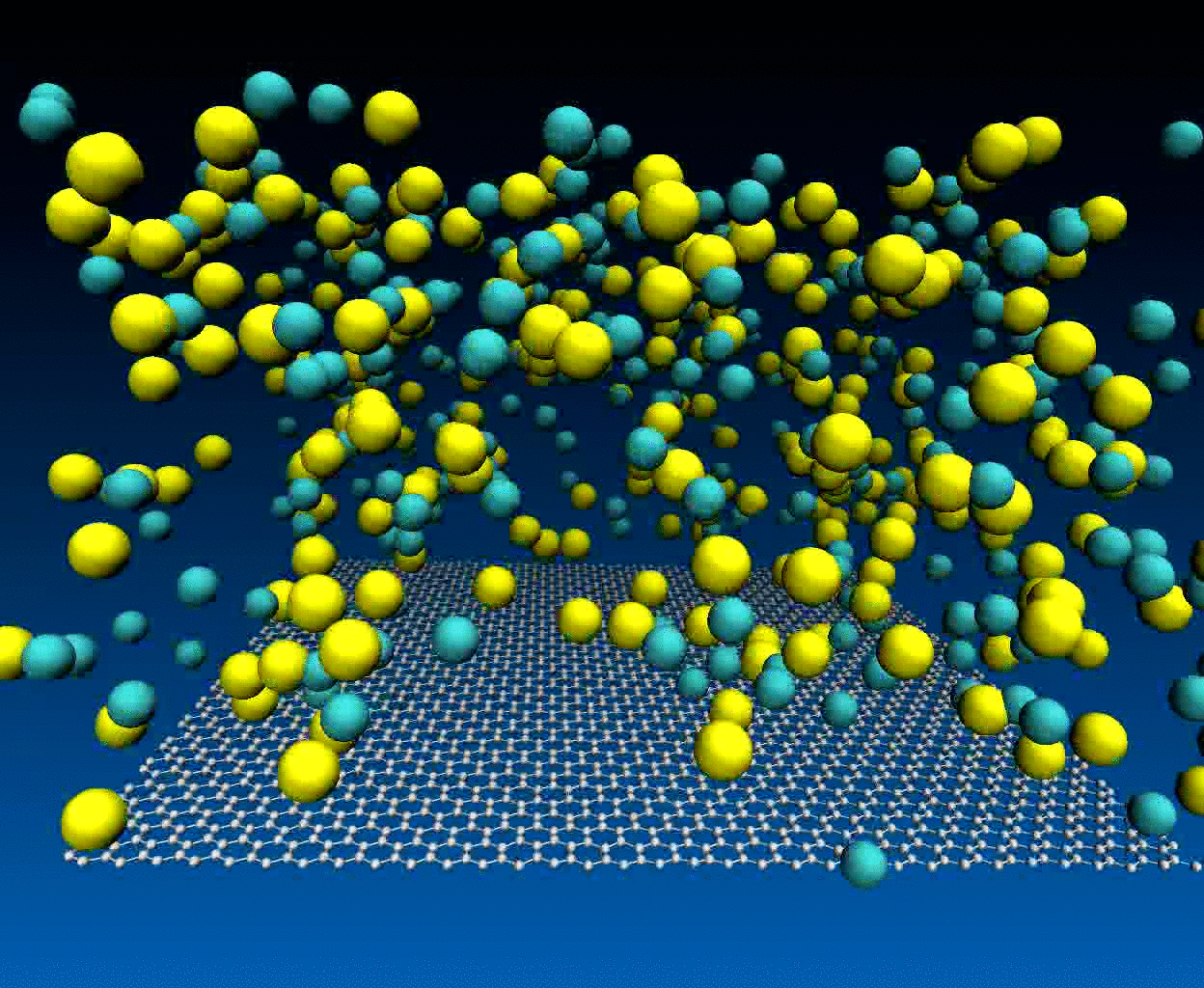
Interactions of NaCl ions with graphene surface
The NaCl interactions with graphene